It’s not just about inbreeding.
When it comes to preserving genetic diversity in a breed: It’s not just about inbreeding.
This statement might shock some new users, and maybe some old. Why, when that’s the primary measurement given on the certificate from UC Davis, would it not just be about inbreeding?
It’s also about allelic richness. What is that? In short, this means lots of different versions of genetics within a closed genepool. Inbreeding systemically over time, throughout a whole population, reduces these versions in number, thereby limiting biodiversity and allelic richness. This is proven. But when we want to stop that loss and maintain what we have, which is the goal of conservation of domestic breeds and species, does breeding solely for heterozygosity (outbred puppies) prevent this?
In this study on Drosophila (yes, fruit flies!) researchers tracked the viability of their specific fruit fly lines by tracking depletion of allelic richness within the line using microsatellites (STRs). Allelic richness, when very low, is an indication that the population is at risk of extinction. Why? Because populations need lots of different genes with their group or species so that they can adapt to environmental and biological challenges.
From the paper:
With careful management, captive populations can maintain high genetic diversity over many generations. However, when population size is small, genetic variation may be lost due to genetic drift. A depletion of genetic variability can increase homozygosity within the population, which in turn may cause lower viability and fecundity (fertility), an effect termed inbreeding depression. Rapid identification of a loss of genetic variation within and/or among populations and generations is crucial.
This is a part of the idea behind the breed assessments created by Dr. Niels Pedersen. What is the state of biodiversity or allelic richness in the breed? As this bank of diversity is depleted, the long term viability of the breed is endangered. This is of utmost concern worldwide in many species and the subject of many papers in recent years.
Why, then, could we not just re-engineer our breeds by just not inbreeding? Wouldn’t ensuring outcrossed offspring also ensure that the breed maintains diversity?
In short, no. Breeding solely for better measures of heterozygosity can reduce breedwide allelic richness over generations, as found in this Master’s thesis based on bison herd management.
This phenomenon is especially exaggerated in breeds with bottlenecks, because they may have only 2-3 alleles per locus (genetic versions) that are very common in the breed and many others are so rare, they are likely to be bred out inadvertently.
Here is an example: Dog A with, say, many particular very common genes could be bred to a dog B with a different set of very common genes, creating measurably outbred puppies. But since both parents have mostly common genes to offer, those puppies are likely to inherit only common genetics, which is then all they have to contribute to the next generation, further breeding toward a bottleneck. Say there was another dog, dog C, with very uncommon genetics, and we breed that dog to dog A, who is also unrelated. This would also produce equally outbred puppies as those from Aand B, but these puppies would have inherited some less typical genetics to contribute to the next generation.
This is why BetterBred offers breeders a way to sort your breedings. A Category 10 breeding alone will ensure outbred puppies, but looking at the breedings only this way may further deplete the population. So we have “potential breedings by outlier index” and “potential breedings by average genetic relatedness.” These two options will help prevent further loss of biodiversity from your breeding population by also putting emphasis on dogs that are genetically unique in your breeding population. Selecting from the top part of these lists, and there is one for every category, will allow you to breed away from any breed bottleneck and help maximize retention of genetic diversity in your breed. Don’t forget to check your breeding summary to see how high your IR (inbreeding) will go – we still want to avoid unknown recessives – and how many puppies will fall below your breed average outlier index (OI). As the OI of a breed drops, we know a breed is breeding toward a genetic bottleneck and loss of breedwide diversity.
The following population illustration will demonstrate this concept!
Let’s look at a pretend population.
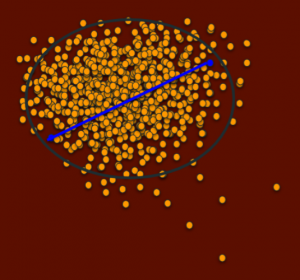
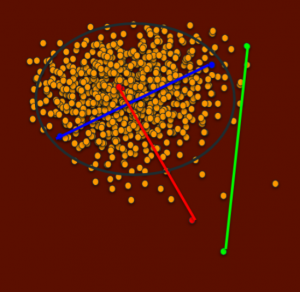
This is a visual representation of the PCoA graphs from UC Davis that represents the population. Each dot is a dog and the distance between them represents how genetically similar or different they are. Dots that are far apart are dogs that are very unrelated and dots that are close together are more related dogs. Notice how this population mostly clusters in one region, with few dogs falling outside of this cluster. Let’s also say this population has a disease that is fixed – meaning the disease genes are in all dogs – in this central population. Dogs outside this cluster will also carry random mutations, since all living creatures do, but they are less likely to be the same mutations as the dogs in the cluster will have, since the cluster dogs are so different from the outside dogs and yet the cluster dogs are all so related, or genetically similar.
Now let’s look at a potential breeding between these blue dogs. The blue line represents how related they are.
These two dogs are distantly related, yet both have genetics that are quite typical for the population.
A breeding between them would have low IRs, but on average more typical genetics for the population. (There will of course be a range since each puppy receives different genetics from its sire and dam).
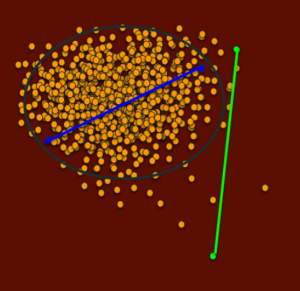
In the next figure, the green dots are also distantly related, and the green line represents how related they are. This line is roughly the same distance as the blue line.
A breeding between the green dot dogs would also be highly heterozygous, seen in low IRs, but will also have on average much less common genetics for the population, because both parents are outside the central cluster of relatedness. (There will of course be a range since each puppy receives different genetics from its sire and dam).
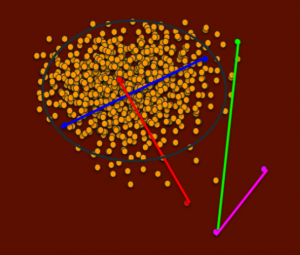
The next set of dogs we will consider is a breeding between a cluster dog with typical genetics and one outside the cluster with atypical genetics for the population.
A dog in the center of the cluster will be highly related to most dogs in the population.
A breeding between the red dot dogs would again produce heterozygous puppies shown in low IRs, but puppies would fall within a range of common to uncommon genetics.
The last pair of dogs we will consider is a breeding between two outlier dogs (magenta line) that are more closely related than the previous potential breedings.
A breeding between them would result in puppies that are somewhat more inbred, shown in higher IRs, but all the puppies would have uncommon genetics. This kind of breeding may be valuable to the breed, because it would increase the presence of the uncommon genetics in the population, which may be on the verge of being lost. Once those genetics are lost, they cannot be reintroduced without an outcross to a similar breed.
The categories on BetterBred are based on the Genetic Relatedness measure between your dog of interest and each other dog of its breed in the database.
Remember to consider the concepts discussed here while using the categories to select breeding mates. Many of the above discussed breedings could be “category 10”, but would also significantly vary in amount of how typical or atypical the genetics produced would be. Outlier Index and Average Genetic Relatedness are the values to look at when considering this.
Breeds that are not yet enrolled need to consider that solely breeding for heterozygous (outbred) puppies is not the answer to the growing issue of genetic diversity loss and rise of complex diseases in dog breeds. More is needed. We are responsible for the future of our breeds.
Remember, it’s not just about inbreeding.

 Previous Post
Previous Post Next Post
Next Post


