Greater Swiss Mountain Dog Preliminary Summary
The Greater Swiss Mountain Dog recently received the preliminary analysis of their breed’s genetic diversity by submitting 34 unrelated Greater Swiss Mountain Dog to UC Davis for analysis. As a result, Dr Felipe Avila has created the initial report. Click here to request GSMDCA to add your Greater Swiss Mountain Dog to the population in our database!
What having more or less genetic diversity means to a breed
After 6 years of carefully analyzing levels of genetic variation and inbreeding in 46 different purebred dog breeds, as well as their health concerns and breeding success, we can see some general trends, trends which are well supported in the large body of scientific literature regarding closed populations and genetic diversity. How healthy and functional a modern purebred dog breed is depends on several factors:
- the number of founders
- the health of those founders
- the historical events or conditions that may have affected the breed
- breeder culture and habits in breeding
By and large, breeds with more diversity tend to be healthier. If a breed’s founders were quite healthy, the breed can remain relatively healthy despite having little diversity if that diversity is not further depleted. Even breeds with a great deal of variation can be largely unhealthy if most of the breed is highly inbred and owe most of their ancestry to unhealthy founders. The healthier breeds tend to have well distributed diversity, meaning each family is genetically different from one another. When a breed is essentially a single family, which is sometimes the case, they are healthier when as outbred as possible.
How inbred is the typical Greater Swiss Mountain Dog?
First off, let’s talk a little about what the researchers at UC Davis are looking at in the report. One aspect of the UC Davis canine genetic diversity test is a calculation called “internal relatedness” that estimates how inbred a dog is. It is widely used by conservation geneticists all over the world. Internal Relatedness, or IR, is a number between -1 and 1. When a dog is more inbred, the number will between 0 and 1; the closer to 1 the more inbred the dog. When a dog is more outbred, the number will be between -1 and 0; the more negative the number the more outbred the dog.
An outbred puppy has inherited markers from its sire that are very different from those it inherited from its dam, so the parents of this puppy are most likely unrelated, or genetically dissimilar. An inbred puppy inherits many of exact same markers from both dam and sire, so the parents of this puppy are most likely closely related to one another, or genetically similar. Being outbred reduces the likelihood of known and unknown recessive genetic diseases being expressed in individual dogs. It also reduces the chances that a complex disease with a recessive component will rear its ugly head. When dogs in a breed are, on average, more outbred, the natural healthy genetic variation in the breed is more easily preserved. When dogs in a breed are, on average, more inbred, the natural healthy genetic variation in the breed is more quickly lost, or bred out inadvertently through a process called genetic drift. All preservation breeders should do their best to reduce genetic drift in their breeds through careful choices of breeding pairs. This loss of existing genetic diversity of a breed, or biodiversity (described simply below), is necessary to maintain their breeds for the future.
Having discussed what the numbers mean above, how inbred or outbred are the individuals in the Greater Swiss Mountain Dog breed?
From the report:
(A)pproximately 25% had IR values
Dr. Felipe Avila
between 0.0882 and 0.3224. Therefore, although the standard genetic assessments made from
allele frequencies indicated that this population of Greater Swiss Mountain Dogs was the product of random mating (Tables 2 and 3), IR values suggest that this assumption is misleading because more outbred dogs are cancelling out more inbred dogs. In reality, approximately one-
fourth of Greater Swiss Mountain Dogs tested were products of closely related parents, some being equivalent to half-siblings or closer.
For perspective, the statement above is unfortunately typical for purebred populations. This means that approximately 75% of the parents of these tested dogs were likely as unrelated as possible within the context of this gene pool; however 25% of the parents were closely related.
While it is clear Greater Swiss Mountain Dog breeders have done a relatively good job producing outbred puppies for a lot of the population, they are either intentionally or unintentionally breeding closely related parents for 1 out of 4 breedings. This is because in purebred breeds with few founders, dogs can seem much less genetically similar than they actually are when looking at even 10 generation pedigrees.
It’s far easier and more accurate to assess whether dogs are unrelated for the breed using genetic testing rather than looking through very deep pedigrees or using pedigree-based calculations. This can eliminate unintentional inbreeding, which happens when pedigrees look unrelated but in fact most of the ancestors farther back than 10 generations are the same ones.
When intentionally linebreeding, genetic testing can allow breeders to assess more accurately how close a linebreeding may actually be, since pairs of dogs with the same pedigree relationship can be more or less genetically similar than one another. This allows breeders to lower the risks of linebreeding while also taking advantage of its benefits to produce predictable puppies. (Remember, though, careful breed management is not just about limiting inbreeding.)
Similarly, breeders can test an imported dog to be sure it is actually an outcross for their breeding program. In many breeds that are popular the world over, this is not always the case. Breeders can benefit from BetterBred’s predictive software to ease the difficulty of selecting less related breeding mates. This can slow the loss of existing variation in the breed as a whole.
After analyzing the existing variation in the initial Greater Swiss Mountain Dog population using a randomly bred “ideal” population of Southeast Asian village dogs (more about this below), Dr. Avila could see from the results that there is very little natural variation left in the breed. He noted, “The curve representing IRVD scores for this cohort (blue line) is shifted well to the right of their actual IR scores (red line), which emphasizes the lack of genetic diversity among Greater Swiss Mountain Dogs when compared to randomly breeding village dogs. Roughly 94% of this cohort (32 out of 34 dogs) have IRVD values of 0.25 or greater (Table 4, Figure 5). This signifies that if they were picked from among from village dogs, all of them would be considered inbred to the level of offspring of full sibling parents or even more closely related than that.”
In other words, if we looked at the tested Greater Swiss Mountain Dog as if they were a part of the naturally breeding village dog population, they would appear much more inbred than they do when compared to their own population’s data. This analysis indicates the gene pool is quite small and likely had a significant bottleneck event. When compared to the canine population with ideal diversity, therefore, they appear to be very closely related. Let’s take a closer look at the genetic diversity, or biodiversity, of the Greater Swiss Mountain Dog breed below.
How much biodiversity exists in the Greater Swiss Mountain Dog today?
Dr. Avila analyzes the data collected from the VGL canine diversity test in different ways in order to assess the total amount of biodiversity within each breed. Many of these measures together give us an overall picture of the state of the breed and as a result we can make informed decisions on breed management.
The best explanation I’ve yet read is from BetterBred Founder Natalie, and as a result we will share it again here!
Most breeders think of DNA as coming in two options- a good gene, or a mutant gene – like it is in many DNA tests. In fact, there can be many versions of each gene or (in this case) marker in a single place on the DNA – like a t-shirt that is available in different colors. The more variants there are, the more information we have about the population genetics of a breed. In more inbred breeds, there are fewer variants for each marker. So to use our analogy, an inbred breed might have only a few colors available in t-shirts, whereas a diverse breed will have t-shirts in many colors. Apart from the relatively small number of genes that make up specific, visible breed traits, the rest of the gene pool is generally healthier when there’s a lot of variation for each marker.
Unfortunately when breeders select too strictly for too long and for very specific traits, there can be an unintended loss of variation in the parts of the DNA that thrive with more variation. A good way to assess whether that good variation has been impacted is by using the markers found in the VGL canine diversity test. Because they are considered neutral – or not associated with any specific known trait – they are great for assessing genetic diversity. In breeds with ample diversity, there will be lots of variations for each marker (lots of colors in the t-shirt drawer.)
But what if you have a breed without much variation? Well, this happens, and can happen quite often. In this case the best thing breeders can do is try to make sure the variants that are in the breed are well distributed – so there are plenty of all of them in the breed. Imagine a t-shirt drawer with lots and lots of red t-shirts but only one blue t-shirt and one green t-shirt. If you lose one of the red ones, it doesn’t change much about the t-shirt drawer – there are lots of other red ones. But if you lose either the blue or green one, the variation is seriously diminished. If, on the other hand a third of the shirts are red, and a third are green and a third are blue, then it’s a lot harder to lose the existing variation in the drawer, even if you lose one once in a while, and even though there are only 3 colors.
The Greater Swiss Mountain Dog’s biodiversity
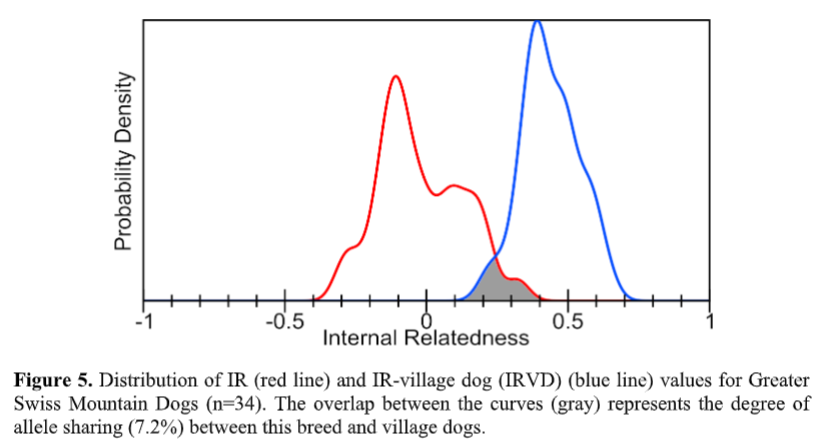
We discussed the individual dogs’ inbreeding levels within the Greater Swiss Mountain Dog population above. However, when we look at the inbreeding level of all individuals in the breed in aggregate – meaning the whole breed altogether- we can gather more information about the breed as a whole. When a whole breed is very inbred on the same few ancestors, it can result in very little remaining genetic variation within the breed. Therefore when the researchers consider their inbreeding as though they were a part of a population of village dogs (often considered “ideal” for biodiversity because they have retained all the genetic variation found now in modern breeds combined, and more), researchers can estimate the amount of biodiversity remaining in a modern breed compared to all the diversity that once existed in dogs prior to breed formation.
The gray area in Figure 5 indicates that Greater Swiss Mountain Dogs tested in this study only
Dr. Felipe Avila
retain 7.2% of the genetic diversity existing in present-day randomly breeding village dogs. This
is similar to Swedish Vallhund (7% retained diversity), another breed with low genetic diversity.
This low amount of retained genetic diversity observed in Greater Swiss Mountain Dogs
becomes apparent when compared to the approximately 30% retained genetic diversity
calculated from comparisons with known alleles at the 33 STR loci of all canids tested at VGL
(section IIB). The Bernese Mountain Dog, another Sennenhund breed previously tested at the
VGL, retains 16.8% of the genetic diversity of village dogs.
This assessment indicates that there has been a significant depletion of genetic diversity in this breed compared to the “ideal”, but let’s look at the data another way:
A secondary way to estimate the loss of biodiversity, due to breed formation and breeding choices post-formation, is to look at the average number of alleles (variations on a single marker) found at each tested locus (location on the DNA strand) and to compare those to the other breeds tested to date by UC Davis.
So, if we look at these numbers what do we find out about this population?
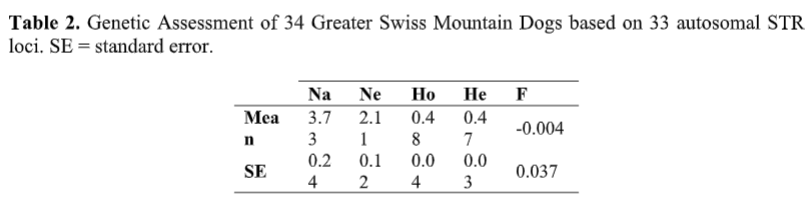
The average number of alleles (Na) identified in this group of Greater Swiss Mountain Dogs was the lowest ever identified for a breed tested at the VGL, corresponding to only approximately 24% of the average number of alleles known to exist at each of these loci across breeds (3.73 out of 15.4 – see section IIA). This number is lower than that found in breeds with low genetic diversity such as the Berger Picard (25%), Polish Lowland Sheepdog (26%), and Irish Wolfhound (27%). When compared to its closely related breeds such as the Bernese Mountain Dog (38%) and Saint Bernard (36%), the lack of genetic diversity found across STR loci in the Greater Swiss Mountain Dog becomes even more apparent. The average number of effective alleles per locus in this cohort was also low (Ne = 2.11). Among the breeds tested at the VGL to date, it was only greater than the Ne value estimated for Berger Picard (1.98).
Dr. Felipe Avila
This number predicts that the Greater Swiss Mountain Dog has retained 24% of the existing biodiversity available to canids; this number is much higher than the estimate from the IRVD, which estimated a retention of only 7.2% of existing biodiversity available to all canines. This can be explained because all purebred canids have lost some biodiversity, whereas Southeast Asian village dogs are likely to have conserved most of the biodiversity that has ever existed in the species.
When looking at the number of alleles found, we consider two things here at BetterBred: the average number of alleles found in total (Na), and the average effective alleles per locus (Ne). The average number of effective alleles are the ones that are effectively contributing to the heterozygosity of a breed. Going back to the t-shirt analogy above, say we have 10 t-shirts, and 8 of them are red and one is green and one is yellow. The total number of shirt colors essentially 3, however the majority color is red, therefore our “effective” t-shirt color would essentially only be 1.
The average alleles per locus in this population was found to be 3.7 (Na). However, Ne (average effective alleles per locus) is a much more telling number, as this is the amount of markers that are effectively contributing to the population’s heterozygosity. This number is currently 2.1 (Ne). Therefore, this report discovered that the breed is on average effectively using about 56.8% of it’s total available biodiversity (Ne/Na). It is possible that this number will increase (or decrease) with more of the population sampled; however, we have found that the health of breeds tends to correlate with how high the Ne/Na ratio is for each breed. This ratio in Greater Swiss Mountain Dog is fairly good, when compared to other purebred breeds, indicating that breeders habitually are not intentionally inbreeding their dogs.
How does a breed community increase the Ne or average effective alleles per locus in a purebred dog breed? They identify individuals who may have unusual genetics for the population, and work to preserve those genetics and redistribute them in the population at large.
Breed relationships
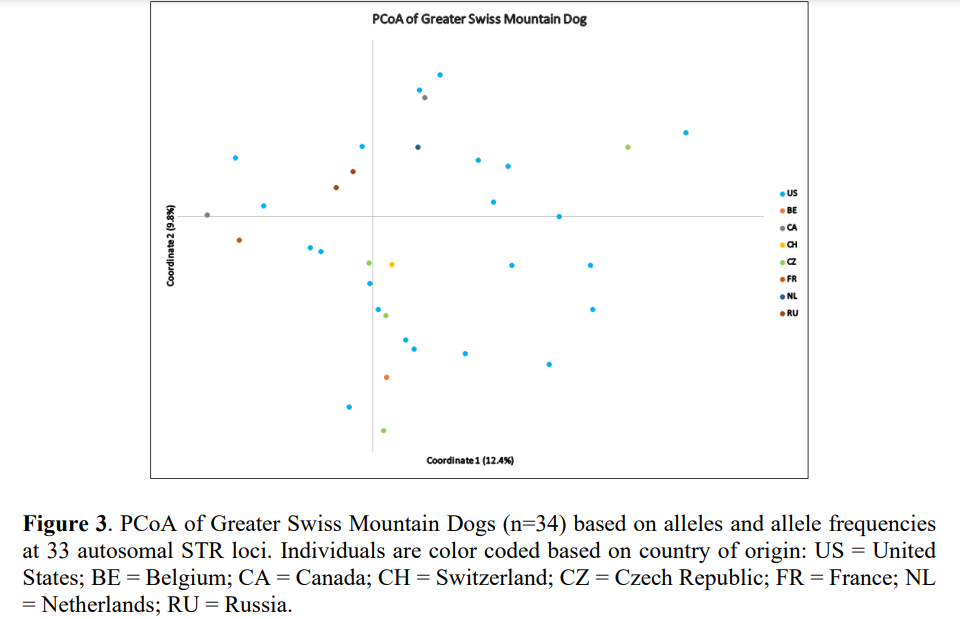
Another use of the data from the VGL canine diversity test is to see how the individuals within the population fit in relation to one another. What do we see when we look at this analysis, called the PCoA graph?
Individual dogs were reasonably dispersed across all four quadrants of the graph with several dogs appearing as outliers (i.e., more genetically diverse), seen on the periphery of the plot (Figure 3). Therefore, it can be assumed that this group of 34 individuals are reasonably unrelated given the low amount of genetic diversity existing the breed. Moreover, individuals from different countries of origin are shown in different colors on the plot; based on the clustering pattern, they form a somewhat genetically homogeneous population, and not genetically distinct varieties based on country of origin. Again, individuals from different countries seen on the periphery of the plot can be considered more genetically diverse (Figure 3).
Dr. Felipe Avila
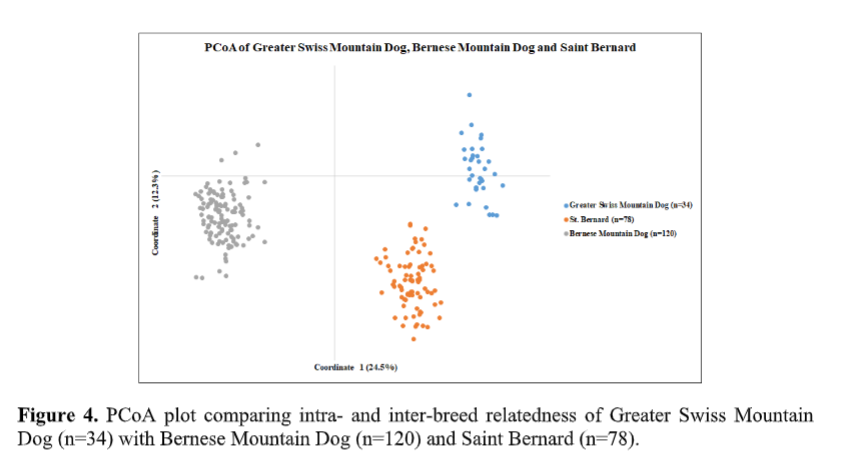
UC Davis VGL also usually looks at how the breed would appear when compared to similar breeds. Here UC Davis compared the Greater Swiss Mountain Dog breed with Bernese Mountain Dogs and Saint Bernards. What did this show?
PCoA shows that the three breeds are genetically distinguishable. Clustering patterns show that
Dr. Felipe Avila
Greater Swiss Mountain Dogs are more closely related to Saint Bernard than to Bernese
Mountain Dogs as the latter breed can be seen further away from the other two breeds, which
cluster closer to each other on the same side of the plot along the X-axis (Figure 4).
The Immune System
The UC Davis VGL canine diversity test can identify more regions of the DLA than any other test available today. The Dog Leukocyte Antigen, called the DLA, is the region of the DNA that codes for the immune system in the dog. There are three regions, Class I, Class II and Class III. This test identifies both Class I and II haplotypes. (A haplotype is a tightly linked group of genes within an organism that is inherited as a group from a single parent.) To learn more about the DLA, read here.
From the report:
Only two DLA class I and two DLA class II haplotypes were identified in this study cohort (Table 5). This number is the lowest found for any breed tested at the VGL, and lower than breeds with equally limited genetic diversity such as the Swedish Vallhund (6 and 4, respectively) and Shiloh Shepherd (7 and 6, respectively). DLA-I haplotype 1012 was hugely predominant, being identified in 94% of the 34 dogs tested; the most frequent DLA-II haplotype was 2003, also at the high frequency of 94%. This suggests a strong genetic and phenotypic influence of a single founder or founder line on both the breed and DLA haplotype diversity. Equal allele frequency distributions identified between DLA-I/DLA-II haplotypes 1012/2003 (94% each) and 1016/2066 (6% each).
Dr. Avila
In summary on the DLA: The Greater Swiss Mountain Dog currently have the lowest amount of variation in the DLA of any breed yet tested at UC Davis, though Bergers Picards had the same low number. Haplotype 1012 paired with 2003 is carried by a large proportion of this population, while the other haplotype is far less well represented. This distribution can be improved by selecting mates with atypical DLA haplotypes and breeding pairs with different Class I/ Class II extended haplotypes. More haplotypes may be found as more Greater Swiss Mountain Dogs are tested.
Conclusions
Based on this report, the Greater Swiss Mountain Dog breed has the lowest amount of genetic diversity of any breed yet tested with UC Davis, though breeders seem to be avoiding intentional inbreeding. Additionally, while some of the breeding pairs are producing relatively outbred puppies, a significant number of dogs are quite inbred, so care must be taken not to cause depletion of the existing diversity in the breed. Because this is the preliminary report, it is hoped that further exploration and expansion of the tested dogs around the globe may reveal more genetic variation existing already in the breed.
Do you own a Greater Swiss Mountain Dog and want to add them to the population? Click here to request to add your Greater Swiss Mountain Dog to the study!

 Previous Post
Previous Post