
Trends in Flat-Coated Retrievers
Last month the Flat-Coated Retriever Society of America held their annual specialty in Oregon and collected an additional 89 samples to test for genetic diversity. BetterBred now has 418 Flat-Coated Retrievers in our database, which is impressive for a lesser known (albeit breathtakingly beautiful!) breed and this large amount of data offers us a lot of power to see what’s happening in a breed using the tools we have on hand.
One of the opportunities we have is to look at the dogs based on age (402 of the dogs have known birth dates) and see if there are any trends happening in the breed. We use our usual well known, peer reviewed population genetics measures to assess trends in genetic diversity in dogs and breeds.
First of all what we know of the breed is that it is likely the result of a genetic bottleneck itself, developed in the same time frame and vein as Golden Retrievers but with a smaller founder base. We can tell that not only by the history of the breed, but also the genetics.
The reason carefully selected markers like those in the Canine Diversity test are good for population genetics is because we can see the more alleles there are at each marker, the more genetic variation remains in the breed.
Most breeders think of DNA as coming in two options- a good gene, or a mutant gene – like it is in many DNA tests. In fact, there are many genes or in this case markers that come in a great many variations – like a t-shirt that is the same exact shirt, but available in different colors. The more variants there are, the more information we have about population genetics. In more inbred breeds, there are fewer variants for each marker. So an inbred breed might have only a few colors in t-shirts available, whereas a diverse breed will have many colors of t-shirts. Apart from the relatively small number of genes that make up breed traits, the rest of the gene pool is generally healthier when there’s lots of variation.
Unfortunately when breeders select too strictly for too long for very specific traits, there can be an unintended loss of variation in the parts of the DNA that thrive with more variation. A good way to assess whether that good variation has been impacted is using the markers found in the VGL canine diversity test. Because they are considered neutral – or not associated with any specific known trait – they are great for a check up on genetic diversity. In breeds with ample diversity, there will be lots of variations for each marker (lots of colors in the t-shirt drawer).
But what if you have a breed without much variation? Well, this happens, and can happen quite often. In this case the best thing breeders can do is try to make sure the variants that are in the breed are well distributed – so there are plenty of all of them in the breed. Imagine a t-shirt drawer with lots and lots of red t-shirts and only one blue one and one green one. If you lose one of the red ones, it doesn’t change much about the t-shirt drawer – there are lots of other red ones. But if you lose either the blue or green one, the variation is seriously diminished. If, on the other hand a third are red, and a third are green and a third are blue, then it’s a lot harder to lose the variation in the drawer, even if you lose one once in a while – even though there are only 3 colors.
In Flat-Coated Retrievers, each marker has an average of 5.79 alleles per locus. This is not very many – compared to the very diverse breed Havanese who have about 9.03 alleles per locus, but it’s more than the Swedish Vallhund, who have about 4.94 per locus. Swedish Vallhunds are a reconstituted breed re-established using just 5 farm dogs, so comparatively this clearly suggests that the Flat-Coats had a small founder base. This also means the breed is relatively highly related – meaning genetically similar – and must be carefully managed to keep the existing diversity – or variation of genes – well distributed so we don’t lose any.
But HOW do we do this? Do we mix and match alleles using our DNA results? Nope. We can’t actually select for any one allele over another because we don’t want to select variants based on numbers. We don’t know what ought to be homozygous or not. We don’t know which ones are rare because they are near bad alleles and which ones are rare just out of happenstance. So we certainly can’t favor any alleles over any others.
So what do we do? We mimic natural processes in two ways: we select for fitness and we maximize genetic diversity. This is how even highly inbred groups of animals in the wild retain more diversity than their pedigrees might suggest.
Natural selection always favors the fittest for the environment, so we too must continue to select for all around robustly healthy dogs with longevity. Breeding dogs should of course pass the breed club’s recommended health testing in each breed and fit the breed standard – but more than that, they should be generally fit – “good doers and easy keepers”. This is the best way to be sure we are selecting very generally for good genes and not for bad ones. And we have to be careful not to be kennel blind in either direction. We don’t want to ignore aesthetic faults, but we also don’t want to preserve fragile but beautiful dogs. We have to make sensible breeding decisions in each generation.
Maximizing diversity is a little trickier for breeders. It is well established that wild canids and other animals instinctively select as mate the least related available, fit mates. Note that I am not saying they never breed to relatives – they do – but they instinctively select the least related available. They do this, it is hypothesized, by pheromones – which are created by the same region of the DNA that controls the immune system. They can smell the more attractive potential mates. (Consider this next time you see a bitch in heat refuse one dog and flirt with another! They may instinctively know something we don’t.)
But since we can’t tell who is least or more related except by pedigree – which isn’t always accurate – we can use genetic relatedness to tell us how similar or dissimilar dogs are. Conservation geneticists have developed very good ways of estimated what is called “pairwise relatedness” – and it’s a complex formula using all the DNA data from panels like the UC Davis canine diversity test. The accuracy depends on how well selected the markers are, how many alleles are found at each, and how related the population is altogether. So far these calculations do an excellent job of identifying relatives accurately – and identifying dogs that are very different from one another. By breeding less related dogs, genetic measures of inbreeding stay low. Overall, FCR breeders have done a good job of keeping inbreeding fairly low. This is a great thing for the breed.
So selecting for health and breeding to the least related available fit dogs is the most basic way to maximize retention of diversity in a breed, but there is one more important aspect breeders must watch out for.
Being unrelated isn’t quite enough information and here’s why. Usually in a breed with bottleneck, there are a great many dogs that are very similar to one another – but there may be a few unusual dogs left that are less related to the bottleneck than most dogs in the breed. We call these genetic outliers. Identifying these dogs is very important, because as long as they are healthy, they are often the last remaining source for the original variations of alleles in the breed. These dogs must be identified, preserved carefully and bred carefully too. They will be unrelated to most dogs in the breed, and as such the temptation will be to breed them to lots of more common mates. However, this only halves their unusual genetics – so while that’s fine to do with caution, it doesn’t have much impact on the breed. It’s like a drop of red paint in a bucket of white. So it’s crucially important that they are bred to other unusual mates so that there is more of the unusual genetics to go around. It’s like increasing that red drop to a cup of red paint – so it has some impact on the bucket of white paint. If outliers aren’t managed carefully, their remaining genetics can be drowned out by the existing bottleneck. If they are managed well, they can improve the distribution of existing variation and help make a healthier breed overall.
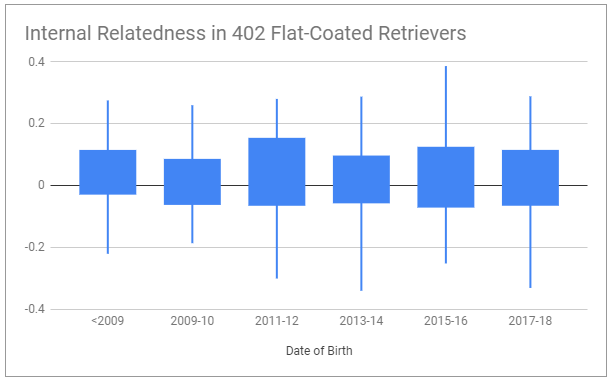
What happens to a breed where these things are not carefully managed? The less genetic diversity a breed has, the more rapidly it is likely to lose variation. Even with breeders selecting for health and using pedigrees or pedigree based COI, breeds can become increasingly related. Overall, Flat-Coat breeders are doing a pretty good job of preventing this – but still the breed is becoming slowly more and more related.
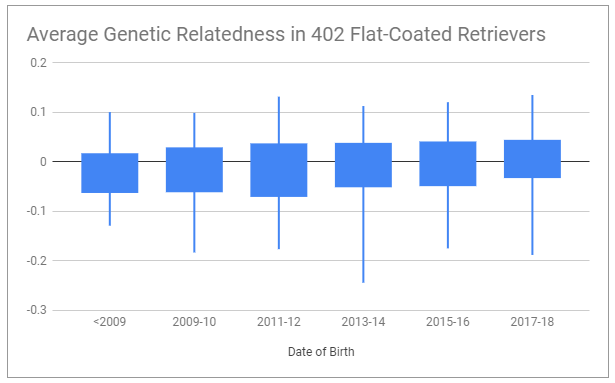
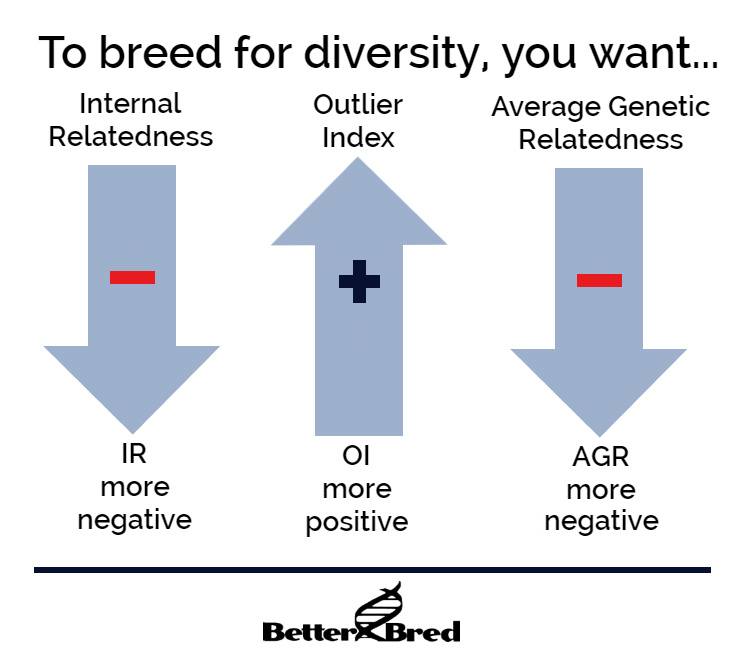
It is possible to reverse this trend. It may seem complicated, but we make it as simple as we can. For this, we use two different measures – average genetic relatedness (AGR), which shows if a dog is highly related to more dogs in the breed, and our own proprietary formula, the outlier index (OI). These two measures use different ways of identifying unusual dogs. Low AGR and high OI mean a dog is unusual. It’s not a good or bad thing – assessing quality means assessing the actual dog. But it’s an important measure for retaining as much genetic variation in the breed as possible, because every healthy outlier is like a time capsule of the original diversity in the breed.
This is why we recommend breeding for lower than breed average AGR, and higher than breed average OI to retain variation, and lower than breed average IR to limit inbreeding. Sometimes we have to breed unusual dogs that might be more related to one another, resulting in higher inbreeding in the line, in order to increase the population of a high quality unusual line. But for the most part, but following these three simple measures – all available for free on BetterBred, a breed community can better preserve their breed for the future.
 Previous Post
Previous Post Next Post
Next Post


