
Initial Breed Summary: Irish Setters
In mid March UC Davis released a preliminary analysis of the Irish Setter breed, based on 50 Irish Setters that were sampled at US dog shows. What did Dr. Niels Pedersen and UC Davis VGL find in regards to the Irish Setter breed?
Inbreeding – how genetically similar are the parents of this tested sample of Irish Setters?
Dr. Pedersen found the following in terms of the inbreeding values of this population:
The entire population is on average slightly inbred with a mean IR score of 0.010 (Table 4, Fig. 3). However, one half of the dogs had IR values from +0.010 to +0.400 and one fourth from +0.388 to +0.600.
Interpretation? A large percentage of the Irish Setter population in the US have high inbreeding levels and could use this genetic diversity testing to reduce their inbreeding. Research on other breeds has shown that the IR of offspring from randomly bred full siblings averages +0.25. IR above that amount suggests that parents are even more similar than typical full siblings.
While this data is representative of show bred, US Irish Setters, samples from other countries and lines bred for performance or other purposes would be needed to determine worldwide inbreeding levels.
What might be one reason that inbreeding values are high in this population? As noted by Dr. Pedersen below, the genetic diversity within the breed in the US appears to be lopsided – meaning that of the alleles tested, 1-2 will be carried by the majority while other alleles are far less typical. Having alleles that are very common and carried by the majority would have the consequence of making a true outcross difficult to find. This also means any breed specific diseases become more and more difficult to avoid. Consequently, a breed with well distributed genetic diversity will also have lower average inbreeding values, as unrelated mates are easier to find. It is our goal at BetterBred to help breeds maintain well distributed diversity or to redistribute it when necessary.
From the report:
Loss of alleles is a common feature of pure breed dogs and reflects the small number of founders existing at the time registries were closed. Not only is the number of founders low, but some founders often contribute more of their genetics to the breed than others. This was demonstrated by the disproportionately high incidence of one or two alleles at each locus (Table 1a). These high incidence alleles have been inherited by descent from founding dogs whose genotypes/phenotypes were highly valued and therefore most conserved. A single allele at four loci (AHT137, AHTk253, FH2001, VGL760) occurred in 88%, 82%, 85% and 82% of the dogs, respectively. These alleles are virtually fixed in Irish setters and have been inherited by descent from a founder or founder line that strongly embodied the breed type.
Breedwide diversity
Biodiversity and allelic richness are terms we try to discuss regularly at BetterBred. Namely, how many different versions of genes or alleles exist in each breed? Breeds with more allelic richness and whose genetics are also well distributed throughout the population tend to be healthier. So what did they find about the Irish Setter’s population?
One of the first observations by Dr. Pedersen is the following:
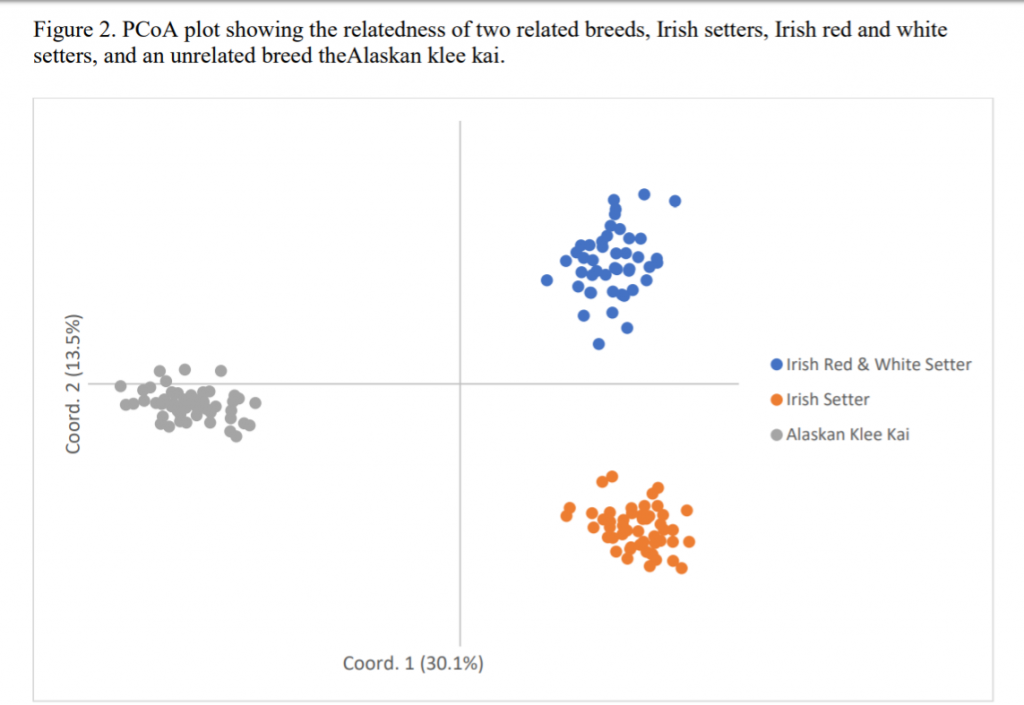
Figure 1 is a PCoA plot of the 50 Irish Setters and 49 Irish Red and White Setters shows them clustering as a distinct breed, as might be expected given differences in founder lines and the many decades that have separated them. Individual Irish Setters are more tightly clustered around the central X/Y axis than individual Irish Red and White Setters, indicating that Irish Setters are more closely related to each other than Irish Red and White Setters.
When Dr. Pedersen added a third breed to the PCoA graph, he concluded that the US Irish Setters sampled for this initial analysis are distantly related to the Irish Red and White Setters thus far sampled (Figure 2).
But what about biodiversity or allelic richness? Irish Setters have low biodiversity in comparison to other tested breeds, the lowest of the breeds tested to date. Breeds with similar low biodiversity include Alaskan Klee Kai, Swedish Vallhund and Shiloh Shepherds – all breeds with very small numbers of known founders. However, as more of this breed population are tested, likely more biodiversity will be found.
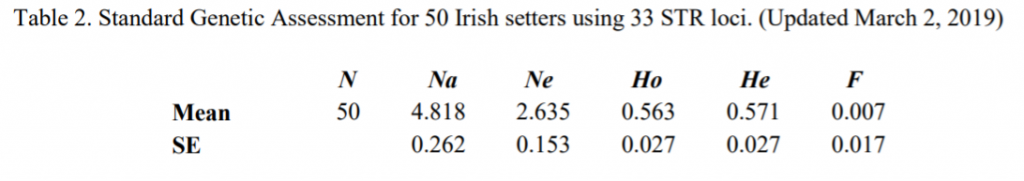
Average alleles per locus and effective alleles per locus are aspects that we discuss in these breed reports. The closer the effective alleles (we call these “alleles that are effectively contributing to your population”) are to the average alleles per locus, the better distributed the genetics in the breed are. Why would you want well distributed genetics? It keeps risk of unwanted recessive traits from appearing, lowers risk of random loss of diversity (called genetic drift) and it lowers the frequency of both simple and complex disease haplotypes that might unintentionally become more common if there were a bottleneck. The average alleles per locus in this breed was found to be 4.818, while the effective alleles per locus was found to be 2.635.
What does this mean? The effective alleles are relatively close to the average alleles per locus than is typical for most breeds, meaning – in this tested sample – the Irish Setter breed community is doing a good job maintaining what they have.
Each breed community should make an effort to raise their effective alleles per locus. How can breeders do this? They should increase the numbers of genetic outliers in the breeding population – which sounds easy, but is hard to do without help, because pedigree based calculations aren’t very accurate at identifying genetic outliers. This is why we developed BetterBred’s breed management software and recommend that breeders select mates and puppies with higher Outlier Index (OI) than the breed average . This measurement was created to help maintain healthy distribution of breed genetics or redistribute unbalanced diversity, and still allows breeders to select for all the traditional traits they want in their dogs.
Diversity compared to village dogs
Dr. Pedersen’s hallmark comparison is to a combined population of village dogs. These domestic dogs have not been shaped by human effort, but instead have evolved through random breeding and natural selection. They therefore have a great amount of genetic diversity which is representative of the original diversity dogs had prior to modern breeding methods. Dr Pedersen calculates the IR results using the breed’s genetic data, and calculates IR based on village dog data, called IRVD, and then compares these two sets of results. The difference between these two results illustrates the diversity that has been lost in each modern breed.
From the report:
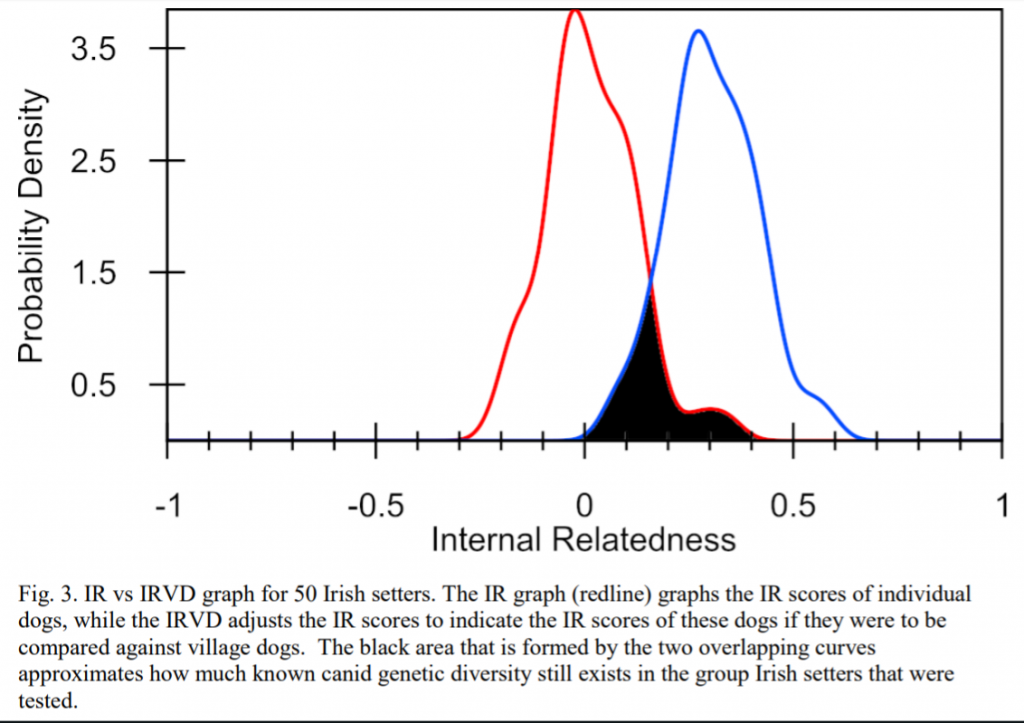
The IRVD curve for Irish setters was shifted well to the right, reflecting an 83% loss (or 17% retention) of potential genetic diversity during breed development (Fig. 3). This 17% estimate of retained genetic diversity is lower than the 30% estimate of retained genetic diversity obtained from known vs. observed alleles and their frequencies presented in Table 1b. This level of 17- 30% retained genetic diversity is lower than genetically diverse breeds such as Toy Poodle (60%), Labrador Retriever (54%), Golden Retrievers (50%), and Alaskan Klee Kai (50%); similar to the Samoyed (35%), Flat Coated Retriever (35.2%), Shiba Inu (29.8%), and Shiloh shepherd (27%); and higher than Japanese Akita (24.4%), Doberman Pinscher (15%) and Swedish Vallhund (7%).
Translation? Irish Setters have lost a significant amount of biodiversity in comparison to the other tested breeds to date. However, this assessment may change as more of the breed is sampled worldwide.
DLA – Dog Leukocyte Antigen
A Look at the Dog’s Immune System!
While only 50 Irish Setters have been tested to date, there is a relatively large number of DLA haplotypes in this breed compared to other breeds with a similar amount of biodiversity.
According to the report:
The Irish setters in this study possessed 8 DLA class I and 9 DLA class II haplotypes (Table 5). Two DLA class I haplotypes (1210, 1211) appear to be unique to Irish setters. The incidence of most of the haplotypes was from 3-18%. However, one DLA class I (1014) and one DLA class II (2037) are found in solely or predominantly in the Irish setter. These two haplotypes are in linkage, forming an even more extended 1014/2037 haplotype that was inherited by descent from an important founder or founder line of the breed.
The report also noted that “The DLA haplotypes 1014/2037 represented 37% of the tested DLA haplotypes, which demonstrates a bottleneck in the DLA of the Irish Setter. Breeders should take note of the atypical DLA haplotypes in the population so as not to lose them to genetic drift. You can read more about using DLA in a breeding program here.
The breed report also noted the following about the two related setter breeds:
Three DLA class I haplotypes and four class II haplotypes are shared between the two breeds (Table 5). However, a larger number of haplotypes were observed in only one breed or the other. It is noteworthy that a founder or founder line with the 1014 DLA class I haplotype played a major role in the evolution of the Irish setter breed, but not in the Irish red and white setter. Conversely, the 1008 DLA class I haplotype was an even more significant contributor to the Irish red and white setters but a minor contributor to the Irish setter.
….
Several of the DLA class I and II haplotypes found in Irish setters were shared with other dog breeds (Table 6). DLA class I and II haplotype sharing were greatest with the Golden retriever, Labrador retriever, and Poodles, as might be expected, given their shared history as outstanding hunting dogs. Significant DLA sharing with the Giant Schnauzer, Havanese and Samoyed were less expected. With the exception of the Havanese, all related breeds had their origins in Europe at around the same period.
DLA comparisons are always extremely interesting as it can inform on origins of breeds; breeds that have more DLA association indicate evolutionary development of breeds, as opposed to those with less DLA haplotypes in common. In this case, the US show lines sampled here further indicate distant relation of the Irish Setter to the Irish Red and White Setter thus far sampled. Irish Setter samples from across the world from conformation and working lines are needed to further study the breed.
Conclusion
From Dr. Pedersen’s report:
IR scores that more accurately measure the relatedness of parents of individual dogs identified a significant sub-population of highly inbred dogs. This might be expected in a small breed with relatively low numbers. Irish setters appear to have below average genetic diversity compared to other breeds, having retained an estimated 30% of
the autosomal STR alleles known to currently exist among all dogs. Based on DLA haplotypes, the breed has evolved around less than 10 founders or founder lines. The lack of genetic diversity in the breed has been confirmed from pedigrees, with an effective population size of 27 distinct individuals (10). There is evidence from this study, as with other reports (4), that breeding for conformation traits leads to more inbreeding and potential loss of breed-wide genetic diversity than selection for performance.
More biodiversity may be discovered as more dogs are tested. Samples are needed from European countries and performance bred lines.
Our Recommendation
The Irish Setters that have thus far been sampled show that there is very little biodiversity within the breed and it will be vital for them to attempt to retain this diversity as the breed is at risk for extinction. More samples are needed to see if more biodiversity can be found, especially if this first initial sample was from a population of related show dogs in the United States.
We recommend that breeders continue to breed to maintain the relatively well distributed diversity of the breed, so as not to lose more allelic richness to genetic drift. Breed for IR at or below zero and OI at or above breed average. When all else is equal, take note of atypical DLA haplotypes and make an effort to redistribute those that are poorly represented. If you have a Irish Setter and would like to contribute to the research phase, you can do so here. Please create an account here, Irish Setter samples from across the world from conformation and working lines are needed to further study the breed.
 Previous Post
Previous Post Next Post
Next Post


