
Preliminary Breed Summary: Irish Wolfhounds!
It’s been a while since we received a new advanced analysis of a breed. Irish Wolfhounds, however, along with several other breeds will shortly be receiving very preliminary analyses of their breed’s genetic state! Breeders will also be receiving their individual reports, which is always exciting and requires some learning to put into use. With their data, they can begin finding the least related available mates and planning their breedings!
So, what did UC Davis’ professor emeritus Dr. Niels Pedersen BS, DVM, PhD find with regard to the Irish Wolfhound in this preliminary analysis? The following article summarizes the full breed report written by Dr. Pedersen. You can find the full report here.
This report is based on testing of 30 carefully selected Irish Wolfhounds; chosen to represent unrelated bloodlines within the breed. While 30 individuals is not enough for a final analysis, it should give a good initial approximation of the status of the breed. We need more Irish Wolfhounds to be contributed, so please consider adding yours!
How inbred is the typical Irish Wolfhound?
A part of the UC Davis canine diversity test is a measurement that estimates how inbred a dog is. This estimate, called Internal Relatedness or IR, is a positive measure from 0 to 1 when a dog is inbred, and a negative measure from 0 to -1 when the dog is outbred. An outbred puppy would inherit genetics from its sire that were very different from those it inherited from its dam. An inbred puppy would inherit many of exact same genetic from both dam and sire. Being outbred reduces the likelihood of known and unidentified recessive component diseases expressing in individuals.
So, how inbred or outbred are the individuals in the Irish Wolfhound breed?
From the preliminary report:
The most outbred dog in the population had an IR score of -0.194, while the most inbred dog in the group had an IR score of 0.321, and the mean (average) IR score for the group was 0.005. The IR curve created from this data was also somewhat triphasic, with one group having IR scores <0.005,a second group with scores 0.005-0.016, and a third group 0.074-0.321(Fig. 3). This latter group contains some individuals that appear more inbred than offspring of full sibling parents. Therefore, IR values give a different picture than that seen with the average scores determined by the standard genetic assessment (Table 2).
Dr. Niels Pedersen 2019
…..(T)he IR scores showed a population of individuals that ranged from reasonably outbred to highly inbred. The inbred dogs are balanced by outbred dogs (…) This is a common feature of all dog breeds.
To translate – some of the breed has very high levels of inbreeding, and because we can’t necessarily detect this without DNA testing and analysis, they could benefit from predictive software aiding in selection of breeding mates, to insure that future breedings are not too close, risking health as a result.
How much biodiversity exists in the Irish Wolfhound today?
Dr. Pedersen’s reports create several different analyses of the data from the VGL canine diversity test in order to assess the total amount of biodiversity within a breed. Many of these measures together give us an overall picture of the state of the breed and as a result we can make informed decisions on breed management.
The best explanation I’ve yet read is from BetterBred Founder Natalie, and as a result we will share it again here!
Most breeders think of DNA as coming in two options- a good gene, or a mutant gene – like it is in many DNA tests. In fact, there are many genes or (in this case) markers that come in a great many variations – like a t-shirt that is available in different colors. The more variants there are, the more information we have about population genetics. In more inbred breeds, there are fewer variants for each marker. So an inbred breed might have only a few colors available in t-shirts, whereas a diverse breed will have many colors of t-shirts. Apart from the relatively small number of genes that make up specific, visible breed traits, the rest of the gene pool is generally healthier when there’s lots of variation.
Unfortunately when breeders select too strictly for too long for very specific traits, there can be an unintended loss of variation in the parts of the DNA that thrive with more variation. A good way to assess whether that good variation has been impacted is using the markers found in the VGL canine diversity test. Because they are considered neutral – or not associated with any specific known trait – they are great for assessing genetic diversity. In breeds with ample diversity, there will be lots of variations for each marker (lots of colors in the t-shirt drawer.)
But what if you have a breed without much variation? Well, this happens, and can happen quite often. In this case the best thing breeders can do is try to make sure the variants that are in the breed are well distributed – so there are plenty of all of them in the breed. Imagine a t-shirt drawer with lots and lots of red t-shirts and only one blue one and one green one. If you lose one of the red ones, it doesn’t change much about the t-shirt drawer – there are lots of other red ones. But if you lose either the blue or green one, the variation is seriously diminished. If, on the other hand a third of the shirts are red, and a third are green and a third are blue, then it’s a lot harder to lose the existing variation in the drawer, even if you lose one once in a while and even though there are only 3 colors.
The Irish Wolfhound’s biodiversity
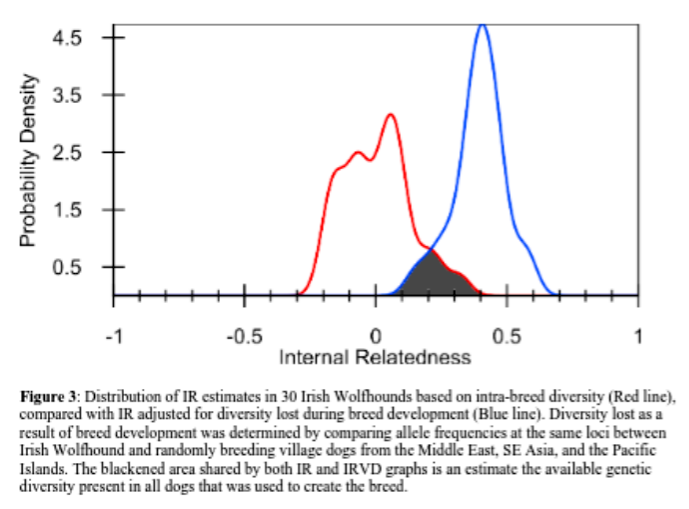
Another use of an individual’s inbreeding value: we can look at the inbreeding level of all individuals in the breed in aggregate – meaning the whole breed altogether. When a whole breed is very inbred, it can mean there is very little biodiversity within the breed. Then, by adjusting those inbreeding values in comparison to the data from village dogs (often considered “ideal” for biodiversity), researchers can estimate the remaining biodiversity of a breed compared to all the diversity that once existed in dog prior to breed formation.
So when this investigation was done, what did Dr. Pedersen see?
The IRVD values emphasize the lack of genetic diversity among the 30 dogs compared to randomly breeding village dogs. If these dogs were picked from among from village dogs, all of them would be deemed inbred to the level of offspring of full sibling parents (IRVD≥0.250). Some would even be equivalent to offspring of village dog parents that were more closely related than siblings(IRVD=0.250-0.600).
…….The IRVD curve for the Irish Wolfhounds tested was shifted well to the right of the IR curve, and the area of overlap was only 13.2% (Fig. 3). It can be estimated, therefore, that contemporary Irish Wolfhounds contain around 13.2% of the genetic diversity that exists in village dogs.
Dr. Niels Pedersen 2019
This assessment indicates very little biodiversity in this breed, but let’s look at the data another way:
A secondary way to estimate the loss of biodiversity within a breed is to look at the average number of alleles found at each tested locus, and compare those to the village dogs. When looking at the number of alleles found, we might consider two things: the average number of alleles found in total (Na), and the average number of alleles that are effectively contributing to the population (Ne – effective alleles). Going back to the t-shirt analogy above, say we have 10 t-shirts, and 8 of them are red and one is green and one is yellow. The total number of shirt colors is 3, however the majority color is red, therefore our “effective” t-shirt color would be only 1.
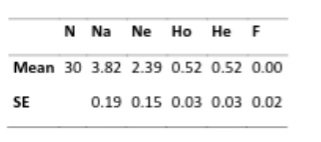
So, if we look at these numbers what do we find out about this population?
“…these 30 dogs retain 3.82/15.4=24.8% of known canid diversity at these loci. However, the average number of alleles is less important than the number of alleles that have the greatest genetic influence on heterozygosity, a figure known as average effective alleles/loci or Ne. The Ne in this group of dogs averaged 2.39 effective alleles per locus. This means that only 2.39/15.4= 15.5% heterozygosity in Irish Wolfhounds was determined by an even smaller portion of available canid diversity.”
Dr. Niels Pedersen 2019
Dr. Pedersen discovered that there is more diversity within the breed than is being effectively used, therefore the estimate of remaining biodiversity is only 15.5%. What do you do in the case of having diversity not contributing to the breed effectively? You identify individuals who may have unusual genetics for the population, and work to preserve those genetics and redistribute them in the population at large.
It is clear from both assessments that the Irish Wolfhound has only a small amount of variation seen so far within the breed. Given that the dogs sampled came from around the world and almost none are closely related, this lack of biodiversity is likely true for the breed as a whole. In fact, only the Swedish Vallhund, a breed restored from only 5 farm dogs, has less diversity. From the report “This level of retained canid genetic diversity is lower than the 60% or so retained diversity observed in the Toy Poodle or 54% in Labrador Retriever, similar to the 15% found in Doberman Pinschers, and higher than the 7% observed in Swedish Vallhund.” These findings may change as more dogs are tested, and it is imperative for conscientious breeders to find and preserve any as yet unseen diversity. How can they do that? First by testing as many dogs as possible, and keeping a keen eye out for healthy unusual or long isolated lines where perhaps there may be more variation.
Breed relationships
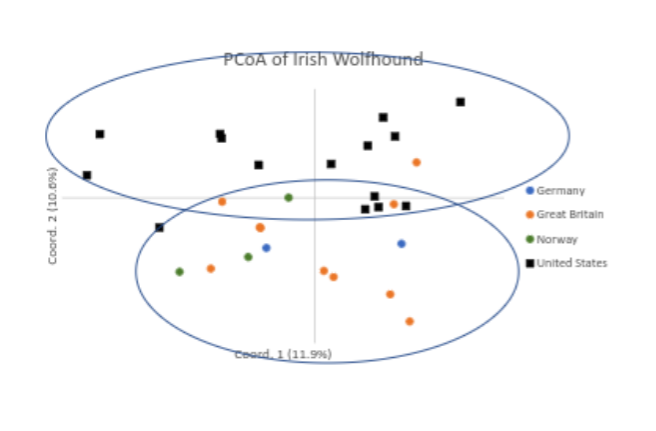
Another use of the data from the VGL canine diversity test is to see how the individuals within the population fit in relation to one another. What do we see when we look at this analysis, called the PCoA graph?
“The 30 Irish Wolfhounds are widely dispersed across all quadrants of the graph, and only two individuals (at 11 o’clock) appear closely related (Fig. 1). Therefore, it can be concluded that these 30 dogs were related to the level of a single breed and carefully selected to represent breed-wide genetic diversity. However, there does appear to be a degree of genetic drift between dogs from the USA and Europe (Fig. 1).”
Dr. Niels Pedersen 2019
Interestingly, it appears that dogs in Europe are clustering away from those in the US. This is not always the case for dog breeds and is good for breeders in each population, because importing dogs may indeed introduce different genetics to compliment breeding programs.
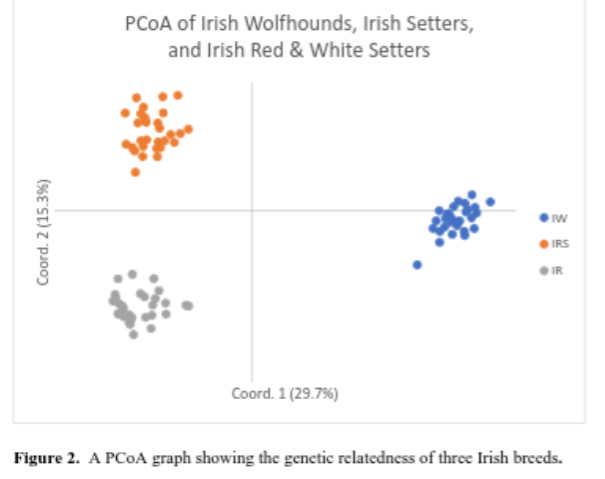
Additionally, the breed’s PCoA can be compared to other breeds. Dr. Pedersen chose to compare the Irish Wolfhound with the Irish Red and White Setter and the Irish Setter. In this comparison, it appears there are individual Irish Wolfhounds that are outliers in comparison to the rest of the breed. It will be interesting to see if these dogs are outliers once run through the BetterBred software program and whether other tested dogs will be found like them.
The immune system
The UC Davis VGL canine diversity test can identify more regions of the DLA than any other test available today. The DLA, called Dog Leukocyte Antigen, is the region of the DNA that codes for the immune system in the dog. There are three regions, Class I, Class II and Class III. This test identifies both Class I and II haplotypes (a haplotype is a tightly linked group of genes within an organism that is inherited as a group from a single parent). To learn more about the DLA, read here.
The number of DLA class I and II haplotypes found in these 30 Irish Wolfhounds was among the lowest of any breed studied to date. The number of DLA class I (n=5) and II (n=4)haplotypes found in Irish Wolfhounds was comparable to Swedish Vallhund (6/4) and Shiloh Shepherd (7/6)
Dr. Niels Pedersen 2019
Unfortunately UC Davis does not have dogs tested from breeds that were used in the reconstitution of the Irish Wolfhound. However, with time it will be interesting to see how the DLA of the Irish Wolfhound compares to related breeds. As stated by Dr. Pedersen in his conclusion below, the lack of biodiversity in the DLA is worrisome. When more dogs are added to the study, there is possibility that more DLA haplotypes will be found.
Conclusions
The following conclusions from Dr. Pedersen sum up the findings from the 30 tested Irish Wolfhounds.
This study confirmed that the Irish Wolfhounds that were tested constitute a single breed, albeit with some genetic variation. Although it is not possible at this point to provide a definitive assessment of genetic diversity, the existing findings are sufficient to make several assumptions. The breed originated from a common founder population that appears to be relatively small, perhaps as few as four lineages, each of which share a common DLA type. It will be important to test more dogs to see if the present findings remain unchanged. If new dogs do not significantly change the present results, it means that the Irish Wolfhoundis the least genetically diverse breed that VGL has studied to date.
Dr. Niels Pedersen 2019
…
Although it appears that contemporary Irish Wolfhoundhave a very narrow genetic base, a lack of genetic diversity is not in itself bad, providing the founder population was relatively free of deleterious genetic traits and breeders have been judicious in avoiding any artificial genetic bottlenecks that may cause either a loss or imbalance of that original diversity. The lack of genetic diversity in the DLA class I and II region of these 30 Irish Wolfhounds is troublesome, but it is uncertain what it means. Certain DLA class I and II haplotypes have been associated with specific autoimmune diseases in certain breeds[25], but autoimmune disorders are presumably not a serious problem in the Irish Wolfhound. Nevertheless, it is important that breeders maintain as much diversity and heterozygosity in the DLA region as possible. Breeds that lack genetic diversity must be managed much more closely to avoid further loss of genetic diversity and have less leeway in dealing with simple recessive or complex polygenic disorders that might arise. Elimination of deleterious traits may result in loss of genetic diversity, especially when diversity is limited.
In a breed with so little genetic diversity at the breedwide level, it will be of great importance to conserve the existing biodiversity so further depletion does not occur due to genetic drift. Breeders can do this by testing their dogs and placing a priority on healthy dogs with less well represented genetics in comparison to the rest of the population, provided they are good representatives of the breed. Using our tools designed to identify these individuals, Average Genetic Relatedness (AGR) and Outlier Index (OI) breeders can allow breeders to select these dogs, as well as to simulate what to expect in potential litters.
Hopefully with further analysis and more dogs submitted to the study, more biodiversity will be found within the breed and successful management and conservation of that existing diversity can begin.
 Previous Post
Previous Post Next Post
Next Post


